Our team focuses on the crosstalk between Ub and UbL proteins in the proteotoxic and genotoxic stress responses. Nevertheless, we collaborate with many public and private laboratories working with distinct biological models or diseases, given the importance of protein modification by distinct members of the ubiquitin family in multiple physiologic or pathologic events.
Protein homeostasis
Protein homeostasis (or proteostasis) is critical to preserve cellular functions. Multiple mechanisms overlap to maintain protein equilibrium and its disruption results in various human pathologies. The regulation of protein stability by proteolysis is crucial to preserve protein localisation and function but also to eliminate proteins that were damaged during their synthesis or after a particular stimuli/stress. Targeting proteolytic pathways is therefore a promising avenue of research in human therapy and in bioengineering.
Proteolysis is regulated in various ways since the cell adapts its activity and response to multiple situations and stimuli. Post-translational modifications play a crucial role in this regulation and include among others phosphorylation, acetylation and various members of the ubiquitin family. One of the most sophisticated mechanisms is the Ubiquitin-Proteasome System (UPS), that uses ubiquitin as a tag to mark the proteins to be degraded by the proteasome. The tagging proteins with Ub can also regulate other functions, such as regulation of signal transduction pathways, enzyme activation, endocytosis, DNA repair, apoptosis, among many other cellular processes. Furthermore, several proteins with the same folding as Ub, called Ub-like proteins (UbL), were also conjugated to other proteins, thereby altering their function or their stability. UbLs also regulate the trafficking of intracellular organelles and the now-called Autophagy-Lysosome System (ALS) that constitutes another major system for intracellular proteolysis.
The Ubiquitin Code
The ubiquitylation of proteins is carried out by an isopeptide bond between the c-terminus of Ub to the Ɛ-amino group of lysine residues on target proteins. Ub can act as a substrate for additional Ub, which are conjugated to any of its seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) or to its N-terminal methionine (M1). The Ub-proteasome system (UPS) uses K48 and K11 polyubiquitylation of substrates to drive their degradation. Ubiquitylation also directs the autophagy-lysosome mediated degradation via K63 chains. Other Ub linkages play very diverse roles including cell signalling, DNA damage response, and protein trafficking. The complexity of this code is increased by mono- and multi-mono-ub modifications, mixed Ub linkage types and other Ub-Like (UbL) molecules such as NEDD8 or SUMO (Figure 2). Researchers now refer to this complexity as the Ubiquitin Code. Deciphering this code and its regulation has become a priority in research and biomedicine, due to its potential to identify specific biomarkers for multiple diseases and new targets for drug development.
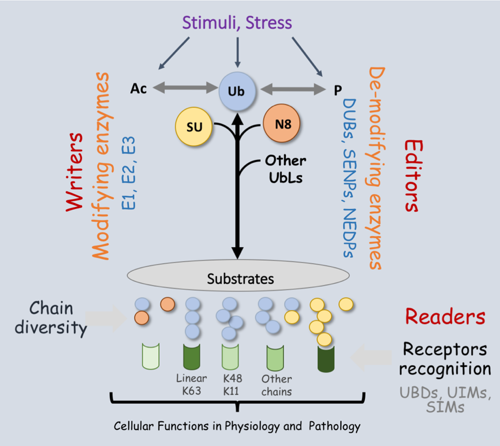
Figure 2 – The regulation of the ubiquitin code.
Each Ub chain architecture is unique and characterized by i) a specific set of enzymes (E1, E2s, and E3s) to generate the chain (writers); ii) a set of deubiquitylases (DUBs) that cleave Ub completely off substrates (erasers) or edit the type and linkage of polyubiquitin (editors); and iii) a variety of proteins containing Ubiquitin binding domains (UBDs) to recognize the chain type(s) (readers) on the substrate to execute the desired effect. Most ubiquitin-binding domains (UBDs) described to this day mainly recognize a specific patch of hydrophobic residues on Ub (L8, I44, and V70) but manage to distinguish between chain types.
SUMO, NEDD8, or other UbLs have parallel enzymatic cascades for conjugation to and deconjugation from substrates. SUMO2/3 paralogs form polymeric chains that are often terminated by SUMO1, which lacks a consensus SUMOylation site. NEDD8 has been shown to form chains in vitro andin vivobut its biologic significance remains to be fully explored. Heterologous Ub/NEDD8 chains are formed in response to stress conditions. SUMO chains are recognized by ubiquitin enzymes such as STUbLs that drive the polyubiquitylation of target proteins. Proteomic studies suggest extensive cross-modifications of unknown function among Ub, NEDD8, SUMO and other UbLs. Therefore, Ub and UbL modifications function as a signaling system with a complex form of regulation. Like ubiquitin, UbLs are signals interpreted by decoder proteins containing motifs/domains that specifically recognize them. The best studied motifs are those interacting with SUMO, known as SUMO-interacting motifs (SIMs)[20]and ATG8. Motifs interacting with NEDD8 and other UbLs exist, but additional efforts are required to fully characterize them and understand their roles in driving specific functions.
Given the complexity of the ubiquitin code, join efforts have been undertaken through several consortia and cooperative programmes (UbiCODE, COST-PROTEOSTASIS, COST-TRANSAUTOPHAGY), in which UbiCARE participates either as a beneficiary or as coordinator.
