UbiCARE develops technology and applications to study the role of several UbLs in distinct physiologic and pathologic events. Our molecular traps to capture proteins modified by Ub and SUMO are already available on the market (TUBEs and SUBEs).
Development of technology and applications to study the role of several UbLs in distinct physiologic and pathologic events
Around 80% of the information available on the proteomes for distinct UbLs is based on the use of tagged molecules to capture modified proteins. This implies that cells must be transiently transfected or infected to generate stable cell lines expressing the tagged forms of UbLs. The formation of particular chains can be compromised by the use of different tags. Independently of this controversial point, cell lines stably expressing these tagged molecules have to be stablished, in the best-case scenario. Transgenic animal models can also be generated to study tissues or organs. However, this strategy cannot be applied to perform studies in humans. We have therefore invested efforts in developing tools that can overcome these technical and ethical issues.
Affinity-based technologies developed by our group have emerged as promising means to identify UbL-modified proteins by mass spectrometry, such as ubiquitin or SUMO traps (TUBEs and SUBEs).Considering their specificity for polyubiquitin chains, UBDs can be used in affinity purification of ubiquitylated proteins. Initial attempts involved full-length ubiquitin-binding proteins. A significant improvement was the multimerization of UBDs coming from various proteins, as is the case with Tandem-repeated Ubiquitin-Binding Entities known as TUBEs (Hjerpe et al).
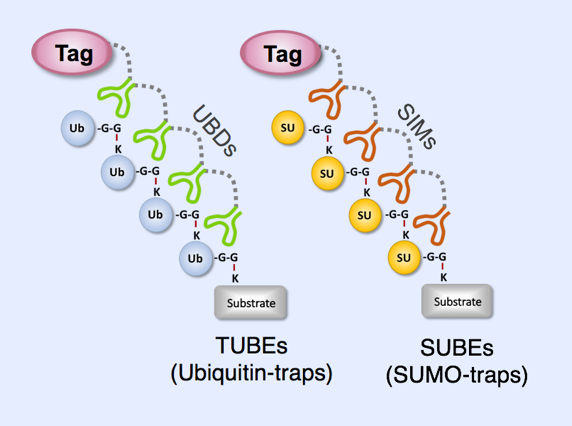
Molecular traps developed by UbiCARE
TUBEs are available on the market and distributed by LifeSensors, testifying to the exploitation potential of the molecular traps that we are developing. Other traps like those capturing SUMO2/3 chains, known as SUBEs, are distributed by SignalChem.
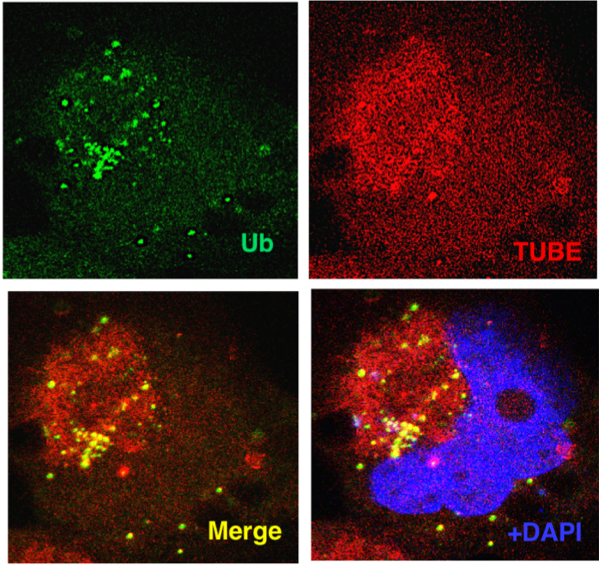
High resolution microscopy using bioTUBEs
The localisation of ubiquitylated proteins was investigated using biotinylated versions of TUBEs, in this case TUBE-Rad23. MCL cells ZBR were treated with BafA and fixed cells were incubated with a Ubiquitin antibody and bioTUBE. A fluorescently labeled secondary antibody and streptavidin were used to detect labelled molecules.
TUBEs
TUBEs consist of tetramerized UBA domains from the proteins UBQLN1 (TUBE1) or HR23A (TUBE2) separated by flexible linkers and fused to multiple tags to facilitate purification and detection. TUBEs bind preferentially to polyubiquitylated proteins, with affinities in the nanomolar Kd range. Their binding also shields the poly-Ub chains from DUB cleavage and from proteasome recognition and degradation.While TUBEs display a preference for poly-Ub chains, they can also be used to isolate highly abundant mono-ubiquitylated substrates. The prototype TUBEs are distributed by the American company LifeSensors.
SUBEs
As for Ubiquitin, similar tandem affinity binding entities have been designed for other UbLs, including SUMO. The four SUMO-Interacting Motifs (SIMs) from the SUMO-dependent E3 ligase RNF4 were used to isolate and identify 300 polySUMO conjugates from cultured eukaryotic cells (Da Silva-Ferrada et al). SUMO binding entities, or SUBEs, have been developed and are similar to TUBEs. SIM2 and SIM3 motifs from RNF4 were tetramerized in tandem and fused to GST, showing that SUBEs interact specifically with polySUMO chains and do not bind to poly-Ub chains or free SUMO moieties. SUBEs prototypes are now distributed by the Canadian company SignalChem. Creating new tools to isolate substrates modified by heterologous or hybrid Ub-UbL chains would perhaps reveal processes regulated in this manner.
Peptides and nanobodies
In addition to the strategies based on existing Ub/UbL-binding domains, other approaches based on novel binding proteins are able to isolate endogenous conjugates. Peptide aptamers, such as affimers, are characterized by small size, a stable folding core domain, and variable domains or peptide loops that can be randomized. Likewise, camelid-derived nanobodies can be derived from in vivosources by immunization or synthetic design. We have developed chain-specific nanobodies using a phage display library in collaboration with the French company Hybrigenics. The first nanobodies we have characterized recognize K48 and K63 chains. These tools are currently distributed by Nanotag biotechnologies. New nanobodies are currently under characterization.
Applications of ubiquitin binding technology
TUBEs and other molecular traps such as SUBEs, alone or in combination with other techniques have been or are currently used for a wide variety of applications. TUBEs are most commonly used as affinity purification reagents for polyubiquitylated proteins. GST or biotinylated TUBEs can either be conjugated to solid supports such as agarose or magnetic beads for affinity purification or be included during the lysis and used as capture reagents. TUBEs can also be associated with immunoprecipitation protocols (TUBE-IP) to identify ubiquitylated forms or interactors of specific proteins such as IkBaor p53.
TUBEs also protect Ub-chains from degradation. By performing TUBEs-based isolation under native conditions, it is then possible to preserve interactors and complexes built around ubiquitylated proteins, such as the proteasome in an example from adriamycin-treated MCF7 cells. Mass spectrometry analysis of TUBEs-enriched extracts identified a unique interplay between the human host and the UPS of P. falciparumover the course of malaria infection (a collaboration with GSK).
TUBEs have been also used in place of antibodies to detect ubiquitylated proteins. Distinct applications were implemented with distinct tagged versions of non-selective TUBEs. For instance, TUBEs labelled with fluorescent tags can be used in co-localizationexperiments in combination with antibodies recognizing specific Ubiquitin-target proteins. TUBEs can similarly be used in far-western immunoblots.
Development of new tools
TUBEs have been incorporated to high-throughput screens (HTS) for monitoring E3 ligase and DUB activities. TR-FRET-based assays monitor autoubiquitylation of E3s, in which biotinylated TUBE is labeled with acceptor fluorophore while E3 antibody is labeled with donor fluorophore. TUBEs have been applied to monitor total cellular ubiquitin in an enzyme-linked immunosorbent assay (ELISA) format, for instance in AlphaLISA or dissociation-enhanced lanthanide fluorescent immunoassay (DELFIA) variations. Multiple detection systems and formats can be set using multi-well plates to search for biomarkers. TUBEs-based protein arrays have been designed for those cases where the amount of starting material is limited, e.g.to detect changes in total protein ubiquitylation after treatment with doxorubicin. Similar applications are currently under development with the distinct technologies developed by our group.
Protein NEDDylation is crucial to regulate protein ubiquitylation due to the direct impact NEDD8 has on the regulation of the Cullin subunits of E3 ubiquitin ligases. Furthermore, under proteotoxic stress conditions many proteins get also NEDDylated. Proteins get NEDDylated trough two pathways. The typical NEDD8 conjugating pathway involves specific NEDD8 conjugation enzymes whereas the atypical NEDDylation involves the E1 of ubiquitin as well. Understanding how these pathways are differentially regulated will be crucial to understand the distinct processes affected by protein NEDDylation under basal or stress conditions. My project aims to identify specific binding domains that can be used to develop NEDD8-binding entities.
The characterization of different NEDD8-binding entities will allow us to use these new tools to decipher protein NEDDylation complexity under different conditions. The biologic significance of normal NEDDylation versus atypical NEDDylation including heteromeric chains of NEDD8/ubiquitin is under extensive investigation in our laboratory. An important role of NEDP1 in the regulation of these chains has been recently uncovered by our collaborators from the CRBM who are also part of the UbiCODE consortium.
